This functions evaluates elastic net repeated cross validation for alpha and
lambda based on glmnet::cv.glmnet().
arcv.glmnet(
x, y,
lambda = NULL,
alpha = seq(0L, 1L, by = 0.1),
nrepcv = 100L, nfolds = 10L, foldid = NULL, balanced = FALSE,
...,
trace.it = interactive()
)
which.min.error(x, s = c("lambda.1se", "lambda.min"), maxnnzero = Inf)
# S3 method for arcv.glmnet
print(x, digits = max(3L, getOption("digits") - 3L), ...)Arguments
- x
arcv.glmnetobject.- y
response as in
cv.glmnet.- lambda
numeric, optional user-supplied lambda sequence; default isNULLandglmnetchooses its own sequence.- alpha
numeric, differentalphavalues that should evaluated (0 = ridge regression, 1 = lasso regression).- nrepcv
integer(1), number of repeated cross-validations (outer loop).- nfolds
integer, number of folds, same as incv.glmnet.- foldid
matrix, an optional matrix withnrepcvrows andnrow(x)columns containing ids from 1 tonfoldsidentifying what fold each observation is in. If givennrepcvandnfoldsare ignored.- balanced
logical, should classes/status be balanced in the folds (default: FALSE)?- ...
further arguments passed to
cv.glmnet.- trace.it
integer, iftrace.it = 1, then a progress bar is displayed.- s
character/numeric, value(s) of the penality parameterlambda. Seeglmnet::predict.cv.glmnet()for details.- maxnnzero
numeric(1), maximum number of allowed non-zero beta coefficients. Default isInfwhich selects the model with the minimal error (the measurement error is chosen from all"lambda.min"or"lambda.1se"results depending ons). If a number is given the model with the lowest (local) error that has at the mostmaxnnzeronon-zero beta coefficents is chosen (also based on the givens, as described above). If no model has less thanmaxnnzerocoefficients the simplest model is chosen and a warning given.- digits
integer(1), number of digits shown in table.
Value
An object of class arcv.glmnet that extends the rcv.glmnet and
cv.glmnet class.
numeric index of model with minimal error.
References
Jerome Friedman, Trevor Hastie, Robert Tibshirani (2010). Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of Statistical Software, 33(1), 1-22. URL https://www.jstatsoft.org/v33/i01/.
Noah Simon, Jerome Friedman, Trevor Hastie, Rob Tibshirani (2011). Regularization Paths for Cox's Proportional Hazards Model via Coordinate Descent. Journal of Statistical Software, 39(5), 1-13. URL https://www.jstatsoft.org/v39/i05/.
See also
Examples
# Examples taken from ?"glmnet::cv.glmnet"
set.seed(1010)
n <- 1000
p <- 100
nzc <- trunc(p/10)
x <- matrix(rnorm(n * p), n, p)
beta <- rnorm(nzc)
fx <- x[, seq(nzc)] %*% beta
eps <- rnorm(n) * 5
y <- drop(fx + eps)
set.seed(1011)
# nrepcv should usually be higher but to keep the runtime of the example low
# we choose 2 here
arcvob <- arcv.glmnet(x, y, alpha = c(0, 0.5, 1), nrepcv = 2, nfolds = 3)
#> Loading required package: foreach
#> Loading required package: future
#>
#> Attaching package: ‘future’
#> The following object is masked from ‘package:survival’:
#>
#> cluster
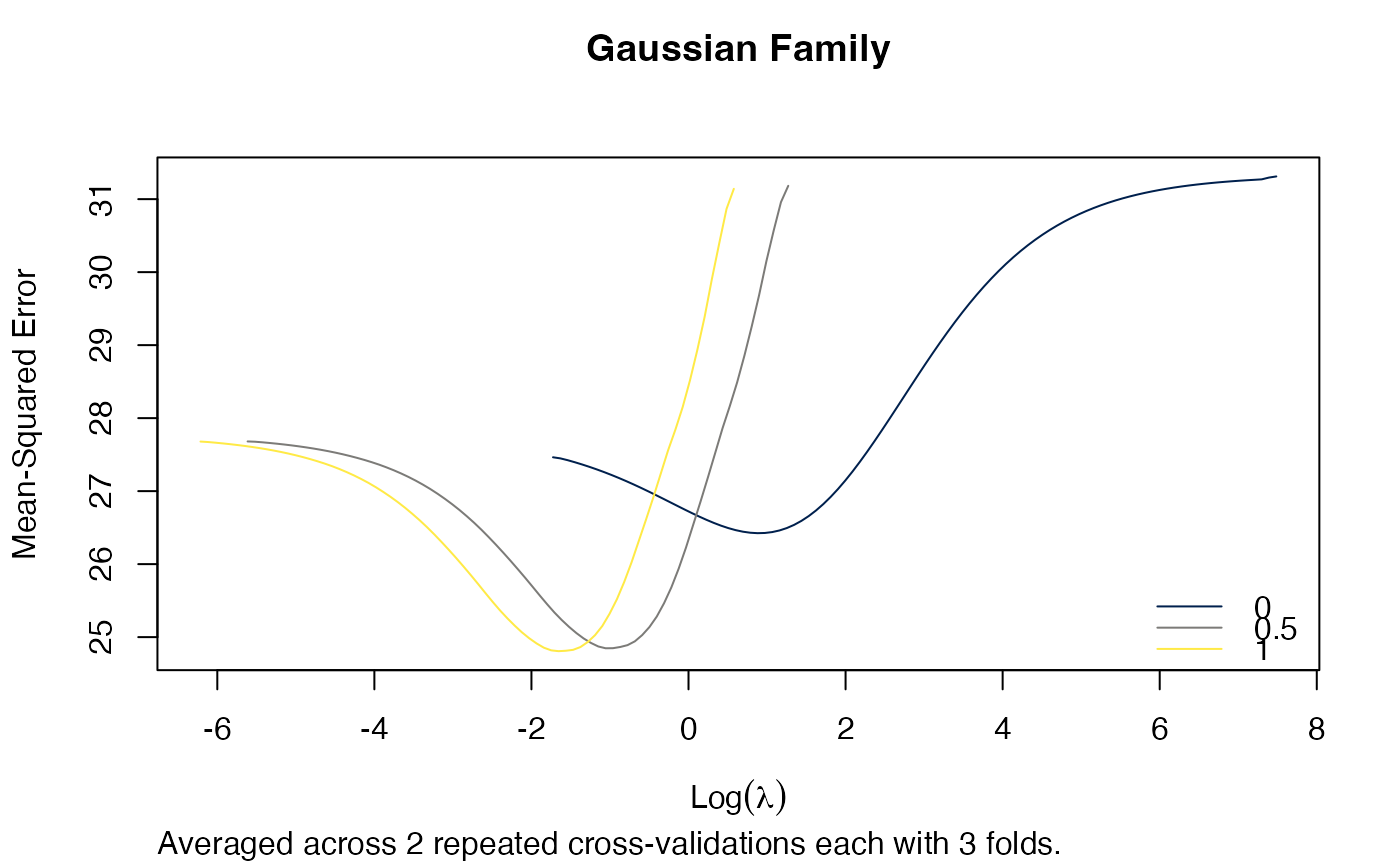
plot(arcvob)
title("Gaussian Family", line = 2.5)
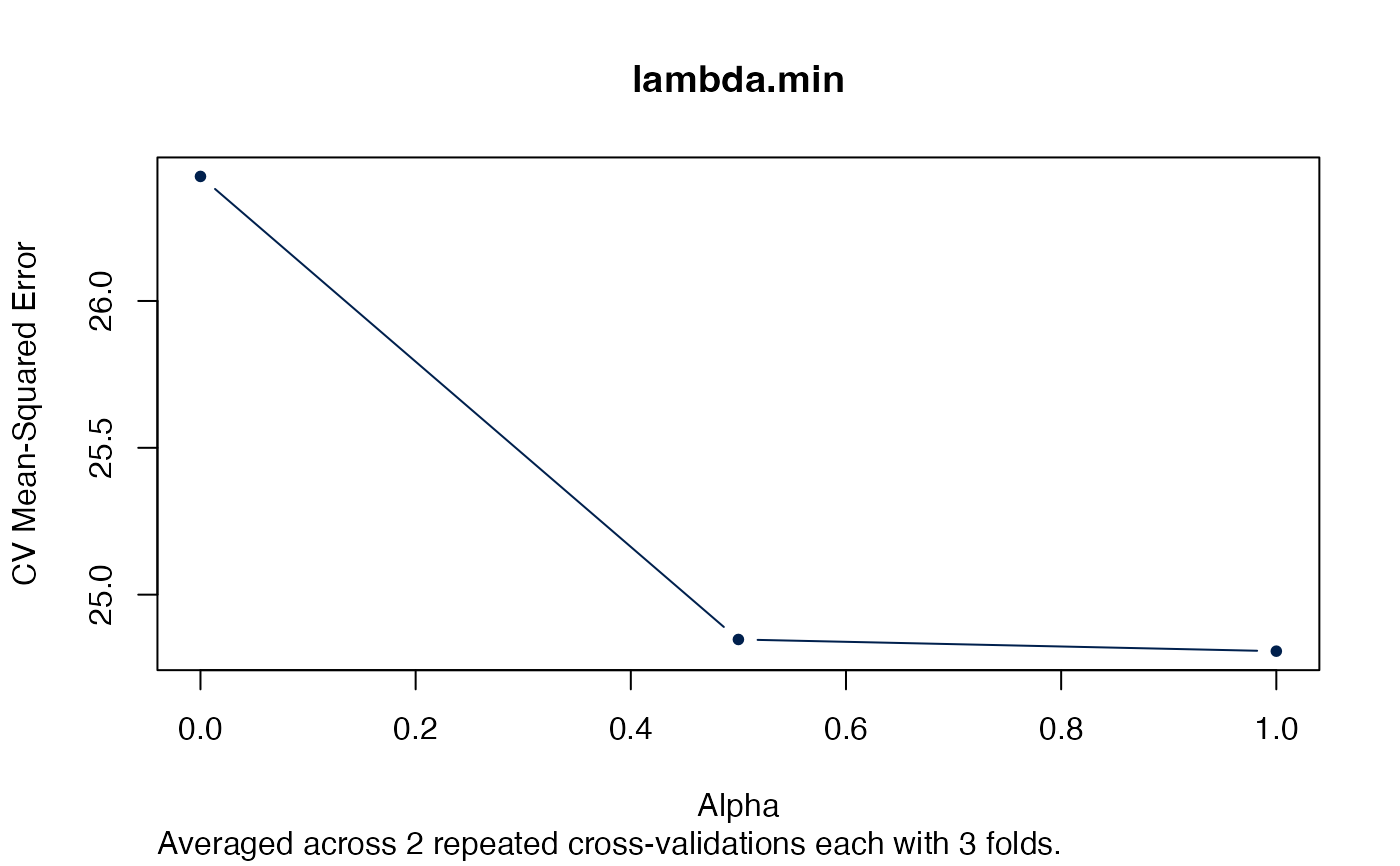
 plot(arcvob, what = "lambda.min")
plot(arcvob, what = "lambda.min")